Quantum Chemistry and Normal Modes of Vibration
Purpose:
The purpose of this experiment is to learn about ab initio quantum chemistry packages and normal modes of small polyatomic molecules.
Equipment and Chemicals:
A PC running Spartan and a printer.
Directions:
See the instructor for directions on how to load and run Spartan.
Calculations:
You will perform an ab initio 3-21G calculation on a small polyatomic molecule.
Your molecule is:
The calculation you will perform may be used to illustrate the connection between charge polarity and the shapes of the frontier molecular orbitals. You will also have the opportunity to caIculate a vibrational spectrum and animate the motions for a few characteristic modes.
- Build your molecule. Select New from the File menu and select the pieces from the model kit to build your molecule. Click on Minimize. In the upper right hand corner you will see information about the minimum geometry of the molecule.
My molecule has symmetry.
Select View from the Build menu to remove the model kit from the screen.
2. Enter the Calculation dialog (Setup menu). Supply a title (e.g., "formaldehyde_321g") and specify Geometry Optimization for Task an 3-21G(*) for Level. Make certain that Charge and Multiplicity are properly set (to 0 and 1, respectively), and click on OK to exit the dialog.
3. Submit the job (Submit under the Setup menu). You will be asked to save the file and to supply a name (e.g., "formaldehyde_321g"). When complete (examine the calculated charges on all of the atoms in the molecule. Select Properties from the Display menu and then Charges from the sub-menu which appears. A message appears below the menu bar.
Charges: Select atoms to display charges.
Click on each atom in the molecule; the atomic charges will be displayed below the menu bar.
E.g., Charges on C2 electrostatic = 0.473, Mulliken = 0.131
Note that the electrostatic and Mulliken atomic charges supplied are different. This reflects the fact that charge cannot be uniquely defined.
The charges on the atoms in my molecule are:
|
Atom |
Electrostatic |
Mulliken |
Focus on the sets of charges on two bonding atoms. Do they confirm your expectations about the polarity of the bonds in the molecule? For example, in formaldehyde, the carbon-oxygen double bond, i.e.,
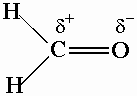
is polar with a partial positive charge (d +) on C, and a partial negative charge (d - ) on O.
When you finish examining atomic charges, click on Done.
Are the atomic charges consistent with the direction of the dipole moment in your molecule? Yes or No?
(To display the dipole moment vector, select Properties from the Display menu and then Dipole from the sub-menu which appears; click on Done when you are finished.)
Draw a picture of the molecule with the dipole vector.
4. Enter the Surfaces dialog (Setup menu). Select HOMO- from the Surface menu. Note that the number "1" appears immediately to the right. This indicates the molecular orbital immediately below the highest occupied "HOMO-1". Click on Add. Next, select LUMO from the Surface menu and again click on Add. Click on OK to exit the dialog. Submit the job (Submit from the Setup menu).
5. When the calculations have completed, select Surfaces from the Display menu. Click on the line "surface=homo{-1}", click on the button to the left of Display Surface and select Solid from among the available display styles. Click on OK to exit the dialog. This will display the Highest Occupied Molecular Orbital (HOMO) molecular orbital on the screen. Print it out and attach to this lab report.
6. Reenter the Surfaces dialog (Display menu). Click again on the line "surface=homo{-1}", and then on the button to the left of Display Surface. This turns off the display of the HOMO. Click on the line "surface=lumo", click on the button to the left of Display Surface, select Solid from among the available display styles. Click on OK to exit Surfaces.
This will display the LUMO of your molecule. Print it out and attach to this lab report. It is generally the case that high-energy filled molecular orbitals corresponding to polar bonds are polarized toward the more electronegative atom, while low-energy unfilled molecular orbitals are polarized toward the more electropositive atom.
7. Reenter the Surfaces dialog (Display menu) one last time to turn off display of the LUMO. Click on the line "surface=lumo", click on the button to the Left of Display Surface, and finally click on OK.
8. Reenter the Calculation dialog and specify Frequency under Task (3-21G(*) should still be selected under Level). Exit the dialog (OK) and then submit the job (Submit under the Setup menu).
9. When the calculation has completed, select Vibrations under the Display menu.
A dialog provides a listing of the vibrational frequencies of your molecule (from smallest to largest) along with their symmetry labels (e.g., A1 or B2). Compare these with the experimental frequencies of your molecule:
|
Mode |
Calculated Frequency |
Experimental Frequency |
Note, that the calculated frequencies are consistently larger than the measured values, typically by about 10%. It is common practice to uniformly scale calculated frequencies by 0.9) to bring them into better accord with experiment.
To animate the individual vibrational motions, click on the frequency in Vibrations dialog, and then click on OK. Experiment with different structure models for best results (ball-and-wire or ball-and-spoke models are usually satisfactory). You should be able to see whether or not the motions you observe are consistent with the experimental assignments.
Attempt to sketch one of the normal modes of the molecule. The following is an example of this type of drawing for one of the normal modes of water:
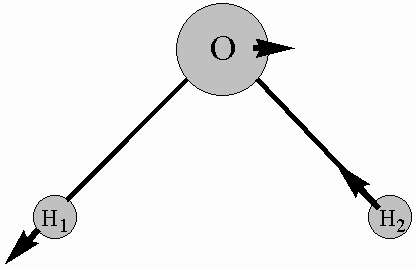
10. Remove your molecule from the screen by selecting Close from the File menu.